Software
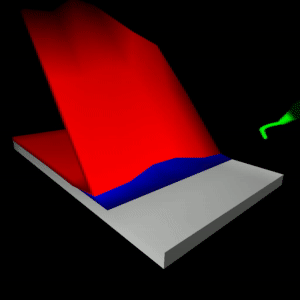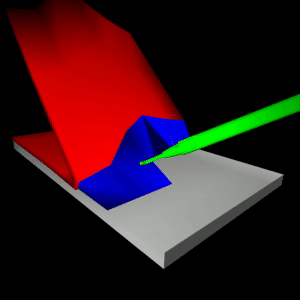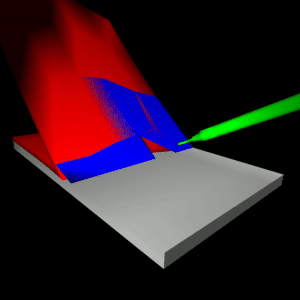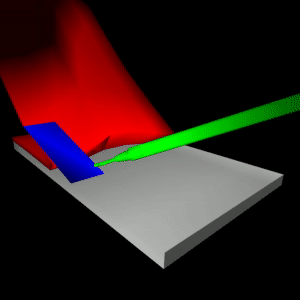
stEVE (simulated EndoVascular Environment)
Framework for the creation of simulations of endovascular interventions using the SOFA framework as backend. Additionally, the following resources are available based on this framework:
- Benchmark implementations of interventions for autonomous endovascular navigation: https://github.com/lkarstensen/stEVE_bench
- Training scripts for RL-based autonomous control in the benchmarks: https://github.com/lkarstensen/stEVE_training
Project paper: https://arxiv.org/abs/2410.01956
Project Repository
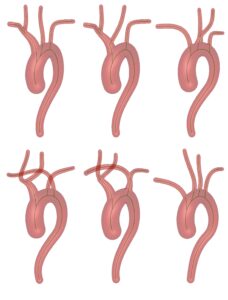
Aortic Arch Generator
A tool to generate artificial aortic arches, e.g., for endovascular simulations. Aortic arches can be utilized as mesh files with full centerline information available.
Project paper: https://doi.org/10.1007/s11548-023-02938-7
LapGym
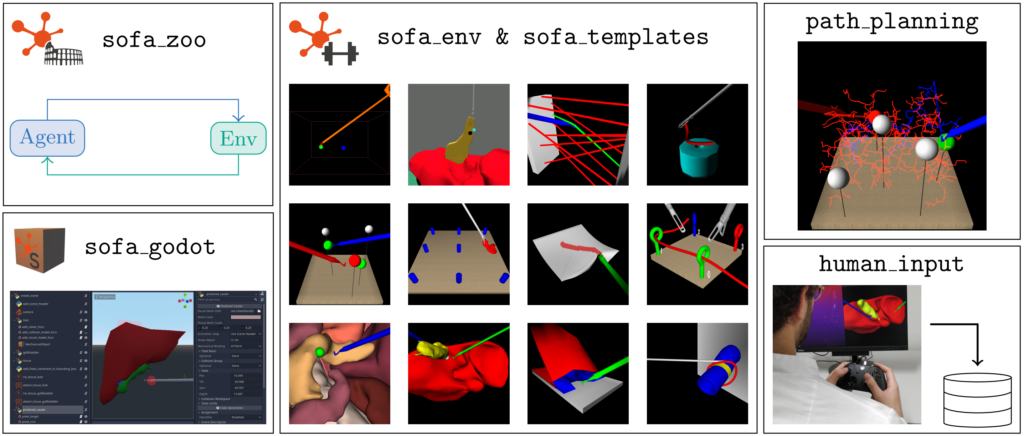 An Open Source Framework for Reinforcement Learning in Robot-Assisted Laparoscopic Surgery
An Open Source Framework for Reinforcement Learning in Robot-Assisted Laparoscopic Surgery
It contains the following sub-projects:
– sofa_env: Defines reinforcement learning environments for robot-assisted surgery using the numerical FEM physics simulation SOFA.
– sofa_godot: Provides a Godot plugin to visually create new SOFA scenes.
– sofa_zoo: Provides the code for the reinforcement learning experiments described in LapGym paper.
Project paper: https://arxiv.org/abs/2302.09606
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Deep Multimodality Image-Guided System for Assisting Neurosurgery
 This project focuses on addressing the challenges in brain tumor surgery, particularly gliomas, one of the most common malignant brain cancers. It emphasizes the need for precise tumor delineation and overcoming issues related to brain deformation during surgery. The project aims to develop an innovative peri-operative image-guided neurosurgery system called DeepIGN. This system involves various components, including a glioma segmentation module, multimodal image registration, explainability module, and an interactive neurosurgical display. These components have been extensively validated in the laboratory and simulated operating room, showing promising results. DeepIGN is designed as open-source research software to advance research in the field of neurosurgery and improve the treatment of brain tumors.
This project focuses on addressing the challenges in brain tumor surgery, particularly gliomas, one of the most common malignant brain cancers. It emphasizes the need for precise tumor delineation and overcoming issues related to brain deformation during surgery. The project aims to develop an innovative peri-operative image-guided neurosurgery system called DeepIGN. This system involves various components, including a glioma segmentation module, multimodal image registration, explainability module, and an interactive neurosurgical display. These components have been extensively validated in the laboratory and simulated operating room, showing promising results. DeepIGN is designed as open-source research software to advance research in the field of neurosurgery and improve the treatment of brain tumors.
Project paper: http://dx.doi.org/10.5445/IR/1000155782
Multimodal CNN Networks for Brain Tumor Segmentation in MRI
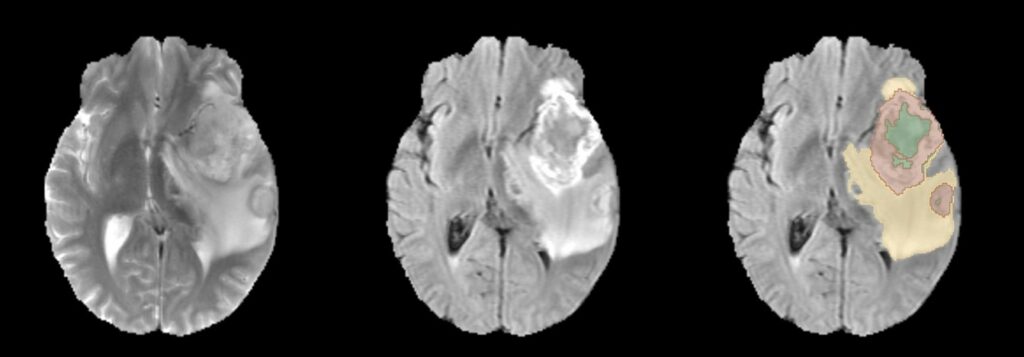
In this project, the focus is on automating the segmentation of gliomas, which is crucial for diagnosing brain tumors, predicting disease progression, and planning patient therapy. The challenge lies in accurately detecting gliomas and their sub-regions in MRI scans, considering variations in scanners and imaging protocols. The project’s contribution to the BraTS 2022 Continuous Evaluation challenge involves an ensemble approach that combines three deep learning frameworks: DeepSeg, nnU-Net, and DeepSCAN. This ensemble achieved top rankings in both the BraTS testing dataset and an additional Sub-Saharan Africa dataset, with high Dice scores and low Hausdorff distances, showcasing its effectiveness in glioma boundary detection.
Project paper: https://doi.org/10.1007/978-3-031-33842-7_11
NeuroXAI: Explainable AI Framework for Brain Imaging Analysis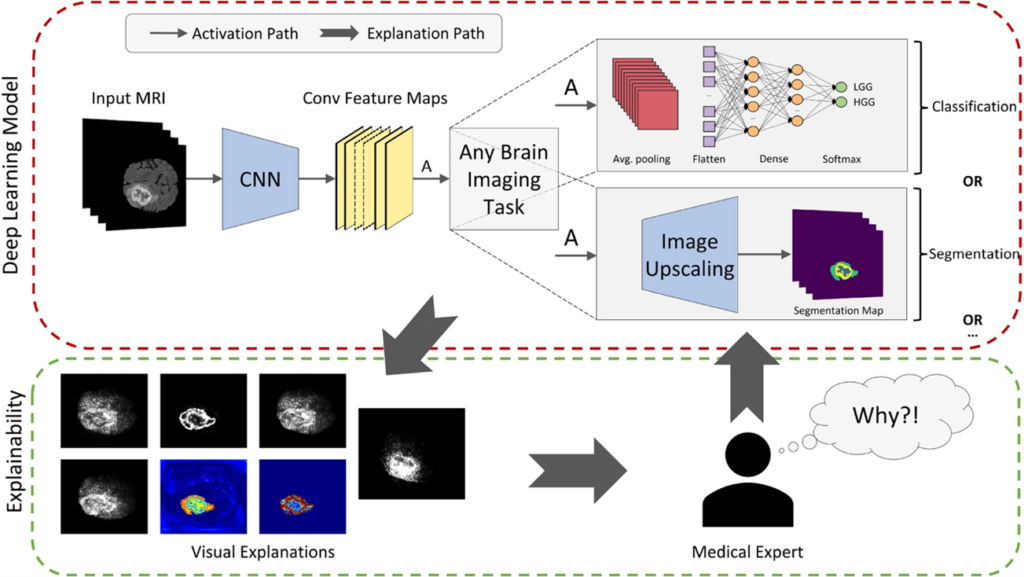
NeuroXAI is a framework for explainable AI of deep learning networks in brain imaging analysis. NeuroXAI allows researchers, developers, and end-users to obtain transparent DL models that can describe their decisions to humans in an understandable manner. This can help medical experts create trust in DL techniques and encourage them to utilize these systems for assisting clinical procedures.
Project paper: https://doi.org/10.1007/s11548-022-02619-x
Slicer-DeepSeg: Open-Source Deep Learning Toolkit for Brain Tumour Segmentation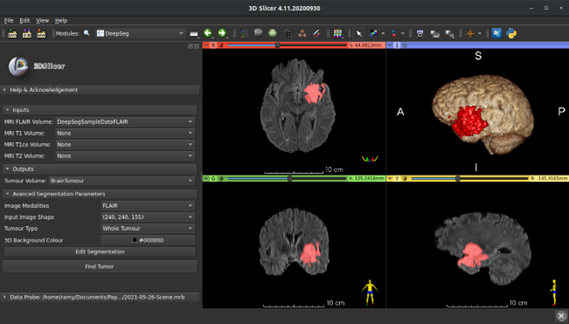
Slicer-DeepSeg is an open-source toolkit for efficient and automatic brain tumor segmentation based on deep learning methodologies for aiding clinical brain research. Pauline Weimann, an MSc student, helped in developing the UI of the Slicer-DeepSeg extension.
Project paper: https://doi.org/10.1515/cdbme-2021-1007
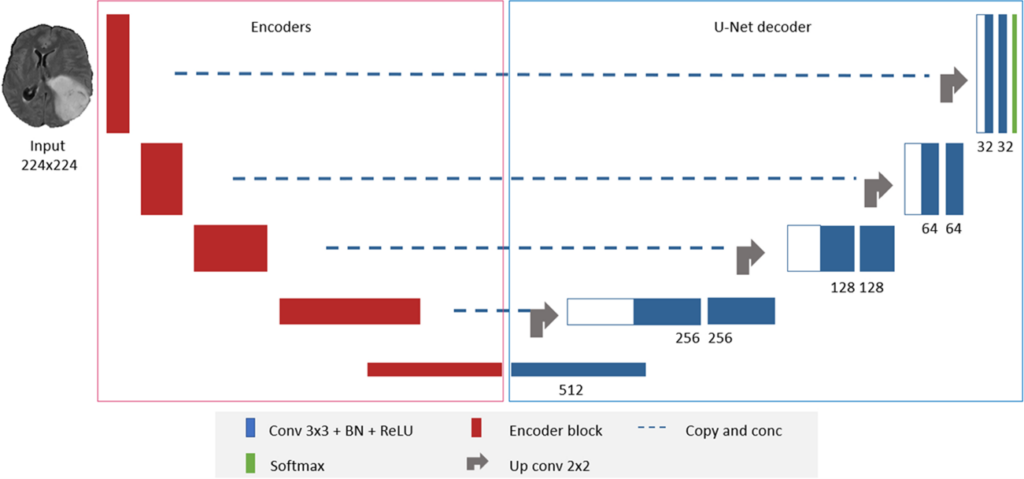
DeepSeg: deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images
DeepSeg is a generic deep learning architecture for fully automated detection and segmentation of brain lesions using FLAIR MRI data. The developed modular framework includes a convolutional neural network (CNN) responsible for spatial information extraction (encoder). The resulting semantic map is inserted into the decoder part to get the full-resolution probability map.
Project paper: http://dx.doi.org/10.1007/s11548-020-02186-z